SNAP¶
Spectral Neighborhood Analysis Potential (SNAP) is a computationally intensive potential in the LAMMPS molecular dynamics software package. The potential computes SNAP force on each atom based on its neighbors and a set of bispectrum components that describe the local neighborhood.
SNAP pseudocode¶
We do not give a detailed descriton of the SNAP potential here but the details of our work is included in our paper on arXiv.
The psuedocode of SNAP is shown here
for(int natom=0; natom<num_atoms; ++natom)
{
// build neighbor-list for each atom
build_neighborlist();
// compute atom specific coefficients
compute_U();
compute_Z();
// For each (atom,neighbor) pair
for(int nbor=0; nbor<num_nbors; ++nbor)
{
compute_dU();
compute_dB();
update_forces();
}
}
The atom loop iterates over each atom, builds its neighborlist based on a cutoff distance. compute_U calculates the expansion coefficients for each neighbor and sums the coefficients for each atom. compute_Z calculates the Clebsh-Gordon product. Then, for each atom, the derivate of atom's expansion coefficients with respect to the position of each neighbor is calculated. Apart from the atom and neighbor loops, each routine additionally has a loop over the bispectrum components.
Background¶
SNAP had a older GPU implementation in Kokkos based on the work done by Christian Trott and Stan Moore which is described at the ECP-copa/ExaMiniMD software repository. The atom loop of the algorithm was distributed across Kokkos teams, which is an abstraction for thread-blocks in CUDA. Hierarchical parallelism was used to distribute the neighbor and bispectrum loops across threads within a threadblock. However the implementation showed a downward trend in its relative performance compared to the peak performance on newer architectures. To address this issue the team joined the NERSC COE hackathon in January 2019 and started collaboration with the NESAP group.
TestSNAP¶
A proxy app called TestSNAP was created as an independent standalone serial computation exactly reproducing the computation shown in the SNAP psuedo-code. The idea was to use the proxy app to rapidly prototype many different optimizations before merging succefull strategies into LAMMPS. It also allowed newer collaborators to rapidly understand and provide useful insights into the algorithm. We encourage the use of proxy apps whenever possible for projects with multiple collaborators since it allows the developers to focus on the core functionality. For the rest of the case study we present the succesfull optimizations on TestSNAP that were merged into LAMMPS. The code for TestSNAP is available here Github.
Refactor¶
One of the biggest disadvantages of the older implementation was the over subscription of limited resources such as registers, leading to a limit on the available occupancy. This was the side effect of a single large kernel launch. To address this issue, the first step was to refactor the SNAP algorithm into individual stages, where each stage can be viewed as a single kernel.
Launching individual kernels allows parameters such as block size to be tailored according to the needs of the kernel. The downside of this is the necessity to store intermedite results of atom and neighbor specific information across the kernels. This adds restrictions on the problem size that can be evaluated on a single GPU since the GPUs have a smaller memory capacity compared to CPUs. To overcome the memory constraint, we used a technique called the "Adjoint Refactorization" which avoided the repeated calculation of the Clebsh-Gordon products. The optimization is explained in great detail in our paper. Shown below is the refactored SNAP code after adjoint refactorization.
for(int natom=0; natom<num_atoms; ++natom)
build_neighborlist();
for(int natom=0; natom<num_atoms; ++natom)
compute_U();
for(int natom=0; natom<num_atoms; ++natom)
compute_Y();
for(int natom=0; natom<num_atoms; ++natom)
for(int nbor=0; nbor<num_nbors; ++nbor)
compute_dU();
for(int natom=0; natom<num_atoms; ++natom)
for(int nbor=0; nbor<num_nbors; ++nbor)
compute_dE();
for(int natom=0; natom<num_atoms; ++natom)
for(int nbor=0; nbor<num_nbors; ++nbor)
update_forces();
Results¶
We present results of SNAP instances that were run on blocks of 2000 atoms with 26 neighbors each on NVIDIA's V100 architecture. Our figure of merit is the time taken to compute the SNAP potential per-atom per-timestep, a metric of throughput which we refer to as grind-time. We call the older GPU implementation on the same V100 GPU as our baseline and compare each of our optimizations against it. For this case study we present the results of ECP's KPP problem, where the algorithm considers 204 bispectrum components. However in most cases that use SNAP potential in LAMMPS, the benchmark problem only considers 55 bispectrum components.
Shown below is the figure of our optimizations compared against the baseline, i.e., the older GPU implementation. The X axis of the figure corresponds to the optimization explained in later sections. The Y-axis corresponds to the speedup over the baseline computed via speedup = baseline / TestSNAP. In this convention, relative speedup of baseline is 1 and a value greater than 1 is an improvement over the baseline. Each successive optimization bar, assumes that all the previous optimizations are in place.
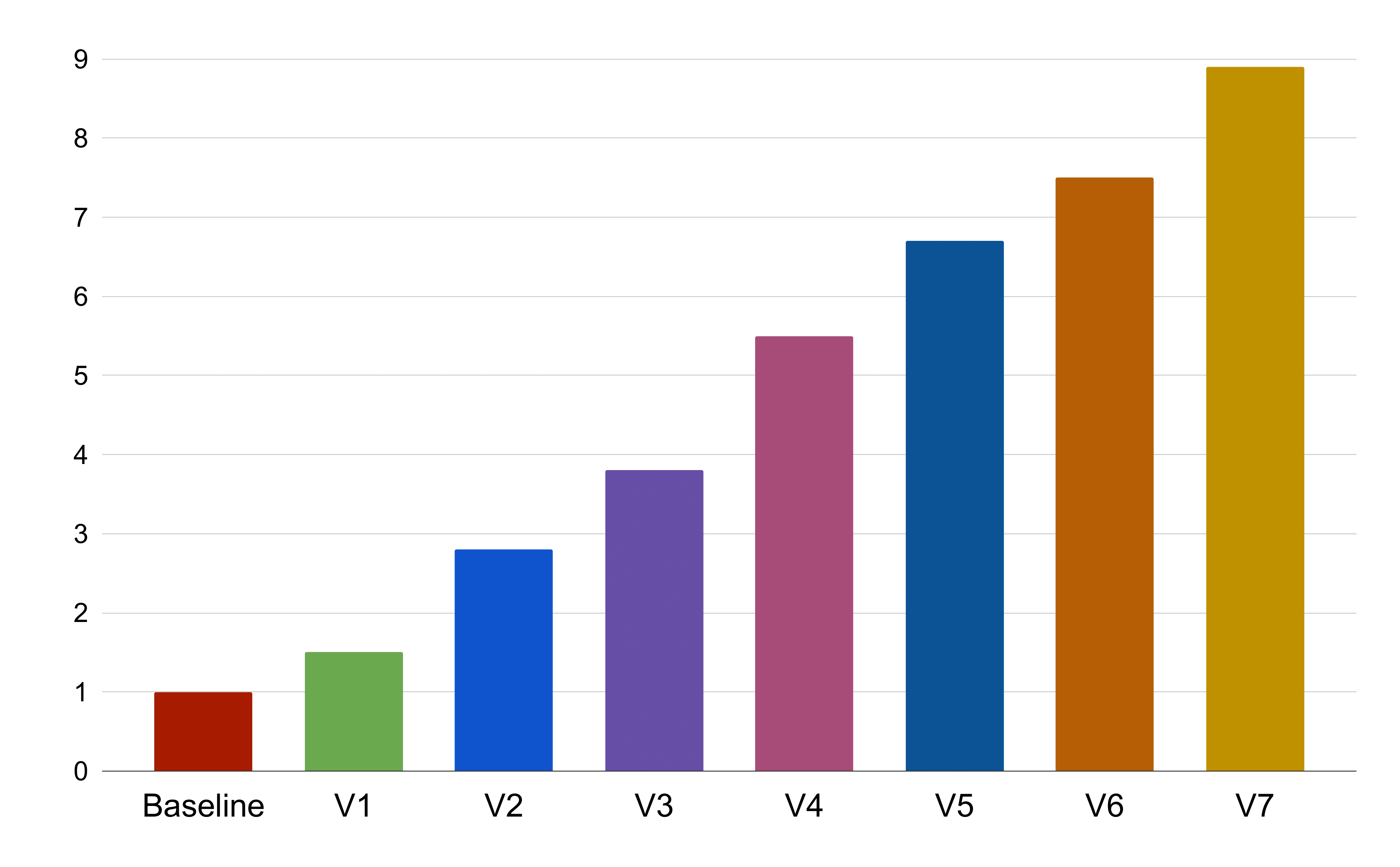
V1 and V2 Atom and Neighbor Loop parallelization¶
The first steps in our paralleization strategy were straightforward where we distributed the atom and neighbor loops across the threads of a GPU. While the atom loop is independent, the parallelization of neighbor loop led to the use of atomic updates in cases where the partial atom specific coefficients over each of its neighbors are accumulated. Although atomic updates are expensive, they allowed us to expose additional parallelism in the code and the trade-off was net beneficial.
At the end of this step, our refactored implementation was ~2x faster compared to the baseline. Even though the baseline implementation also exposed atom and neighbor loop parallelism, our combination of kernel refactor and adjoint refactorization allowed much better GPU utilization and hence a higher throughput.
V3 and V4 Data Layout optimization¶
Unlike CPUs which prefer row major data layout to avoid false sharing and cache thrashing, GPUs prefer column major data layout to promote memory coalsceing. Memory coalescing implies that consecutive threads access consecutive memory locations. Such a data layout is highly efficient on SIMD architectures such as GPUs. In the original TestSNAP implementation, the data structures followed a CPU friendly data access pattern where the bispectrum indexes were the fastest. Switching them to GPU friendly layout where the atom index is faster gave us a 1.3x speedup over the previous optimization.
While changing the data access pattern gave perfect coalescing in routines that only had atom loop, in order to get perfect coalescing in routines with both atom and neighbor loops, the order of the loops were reversed. This was an extremely easy step due to the code refactoring. At the end of V3 and V4 optimizations, the new implementation gained a 4x speedup compared to the baseline.
V5 - Collapse bispectrum loop¶
As mentioned earlier, apart from the atom and neighbor loops, each routine has a bispectrum loop. In our trials, collapsing the bispectrum loop in compute_Y, which is the most expensive routine in the algorithm, gave us a 3x performance boost in the kernel and a 80% performance overall.
V6 - Transpose¶
Another profiler driven optimization was the transpose of one of the data structures between compute_U and compute_Y. Atomic updates to that data structure in compute_U required column major data layout, while due to strided indexing into the same structure in compute_Y required row major data layout for performance. In order to get the best of both, a simple transpose of the data structure was performed between the two kernels. The transpose added a 0.2% overhead but gave a 20% net performance boost.
V7 - 128 bit load/store¶
SNAP performs complex double arithemetic operations. Due to the lack of native complex datatype in C++, a small struct of two doubles was created to represent the complex number. Profiler driven analysis showed that the structure did not make use of 128 bit load/store operations available on NVIDIA V100 architecture but instead generated two distinct 64 bit load/store instructions. Hence in order to assist the compiler, C++ alignas(16) specifier was added to the structure. This gave a 15% performance boost over the earlier existing optimizations.
Architecture specific optimizations¶
All the above optimizations were first tested in TestSNAP and later merged into LAMMPS. Additional architecture specific optimizations such as the use of scratch memory, which is the Kokkos abstraction for low latency shared memory and other algorithmic improvements were directly introduced into the mainline LAMMPS-SNAP implementation. This led to a total of 22x speedup of the latest implementation over the baseline. A deeper discussion on all these optimizations and algorithmic improvements is available in our paper on arXiv.
Lessons Learned¶
- Data Layout is important for optimal performance.
- Coalesced memory accesses on GPUs lead to performace benefits.
- Increasing the AI on GPUs can break the memory bandwidth barrier.
- Profiling can guide optimizations.
- Performance improvements came from algorithmic optimizations and implementation targetting specific hardware.