Running Python on AMD Hardware¶
Perlmutter compute and login nodes have AMD CPUs. Previous NERSC systems such as Cori had Intel CPUs. For the most part, Python users do not have to worry about this difference.
If you're using NumPy with Intel's Math Kernel Library (MKL) backend then you may be missing out on performance optimizations which are not enabled by default on AMD CPUs. The rest of this page will cover details about using NumPy with MKL on AMD CPUs at NERSC.
tl;dr: Try your code with and without fask-mkl-amd module.
module load fast-mkl-amd
How can you tell if you're using NumPy with MKL?¶
Look for "mkl" in the output of conda list:
> conda list
...
blas 1.0 mkl
...
mkl 2021.4.0 h06a4308_640
...
If you installed numpy from the defaults conda channel then you will probably have MKL in your conda environment. If you installed numpy from the conda-forge conda channel then you will probably have an OpenBLAS backend.
You can install numpy with an MKL backend from conda-forge with the following command:
> conda install -c conda-forge numpy "libblas=*=*mkl"
Should you be using NumPy with MKL?¶
Many computationally expensive functions (like those in numpy.linalg) are using optimized libraries like Intel's Math Kernel Library (MKL) or OpenBLAS under the hood. In the past, our advice to NERSC users was generally to use MKL as it was well-adapted for our Intel hardware. On Perlmutter however the CPUs are AMD, so does this recommendation still hold? The answer is yes, but with some important caveats. Spoiler: the best way to know for sure is to benchmark your code.
Intel MKL will check the CPU manufacturer and choose a code path accordingly. This is discussed in detail in a blog post by Daniel de Kok and in another blog post by Donald Kinghorn. Without any intervention, MKL may use a less-optimized code path on AMD hardware.
To use Daniel de Kok's suggested workaround on Perlmutter, module load fast-mkl-amd. For more information about why this matters, please see the results of our benchmarking study below.
Benchmarking study¶
We performed a small benchmarking study on AMD Rome hardware (AMD EPYC 7702 64-Core Processor) to try to estimate Python performance on Perlmutter. You will find information about our benchmarking study, including all of the materials you need should you wish to reproduce it or run it elsewhere, at the NERSC Python AMD Benchmarking Gitlab repository.
We tested the following:
- 4 NumPy functions (
numpy.linalg.eigh,numpy.linalg.cholesky,numpy.linalg.svd,numpy.dot) at - 3 square matrix sizes for
- 3 library configurations (
mkl,mkl-workaround,OpenBLAS) for - 2 different configurations (high arithmetic intensity,
highAI, and high memory bandwidth,highMB) - Using 1 AMD EPYC 7702 (Rome) with 64 physical cores and 2 hyperthreads per core
We determined the high arithmetic configuration using the standard DGEMM matrix multiplication benchmark; you can find more information about this benchmark at the Intel GEMM benchmarking page. We adjusted the thread and process binding on the AMD Rome to optimize DGEMM GFLOPS/s using the following configuration:
OMP_NUM_THREADS=128 OMP_PLACES=threads OMP_PROC_BIND=close srun -u -n 1 -c 128 --cpu_bind=sockets ./dgemm.exe
We determined the high memory bandwidth using the standard Stream-Triad benchmark; you can find more information about this benchmark at the Intel Stream-Triad Memory Bandwidth Optimization page. We adjusted the thread and process binding on the AMD Rome to optimize Stream-Triad GB/s bandwidth using the following configuration:
OMP_NUM_THREADS=64 OMP_PLACES=cores OMP_PROC_BIND=spread srun -u -n 1 -c 128 --cpu_bind=sockets ./stream-triad.exe
This is not a perfect test, nor is it exhaustive. Our goal is to understand general NumPy performance on AMD Rome so we can provide this information to our users to make informed decisions about which libraries to use on Perlmutter.
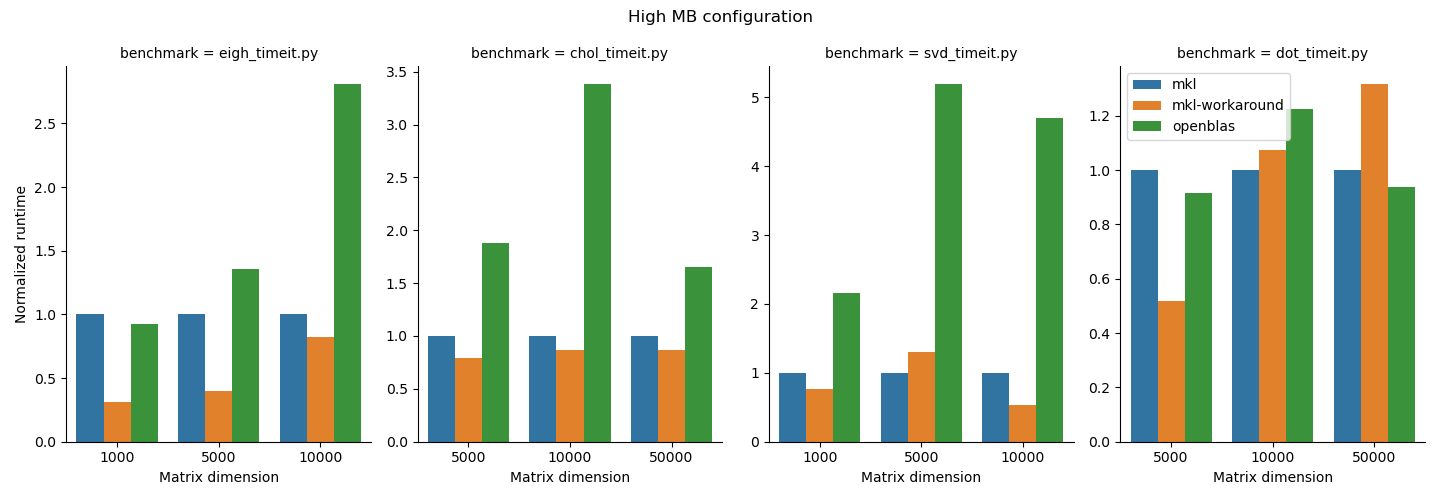
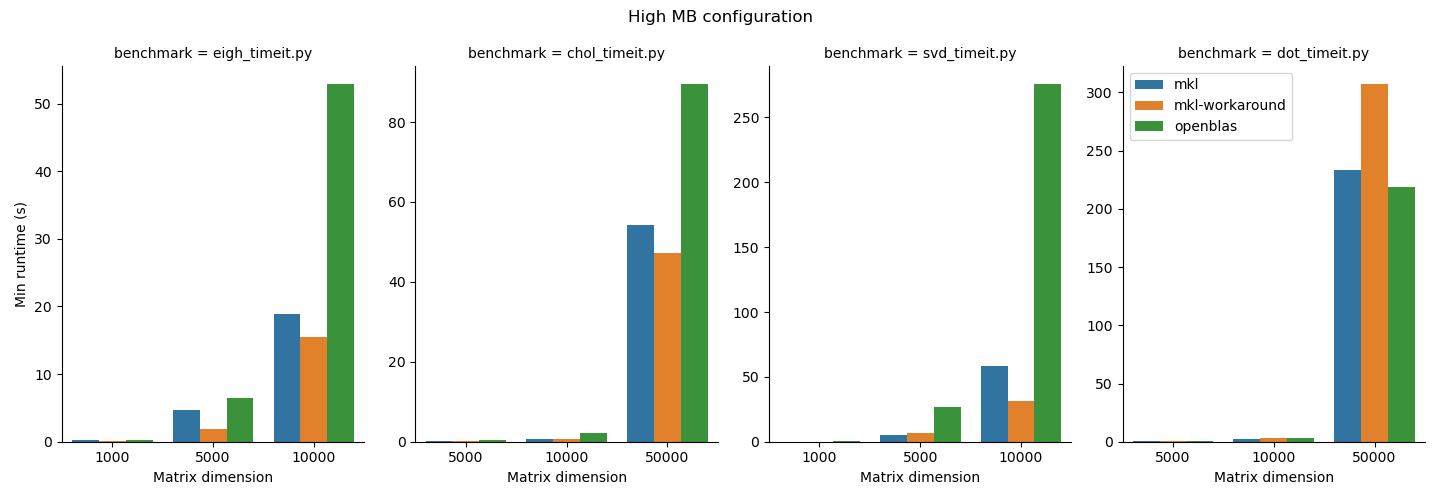
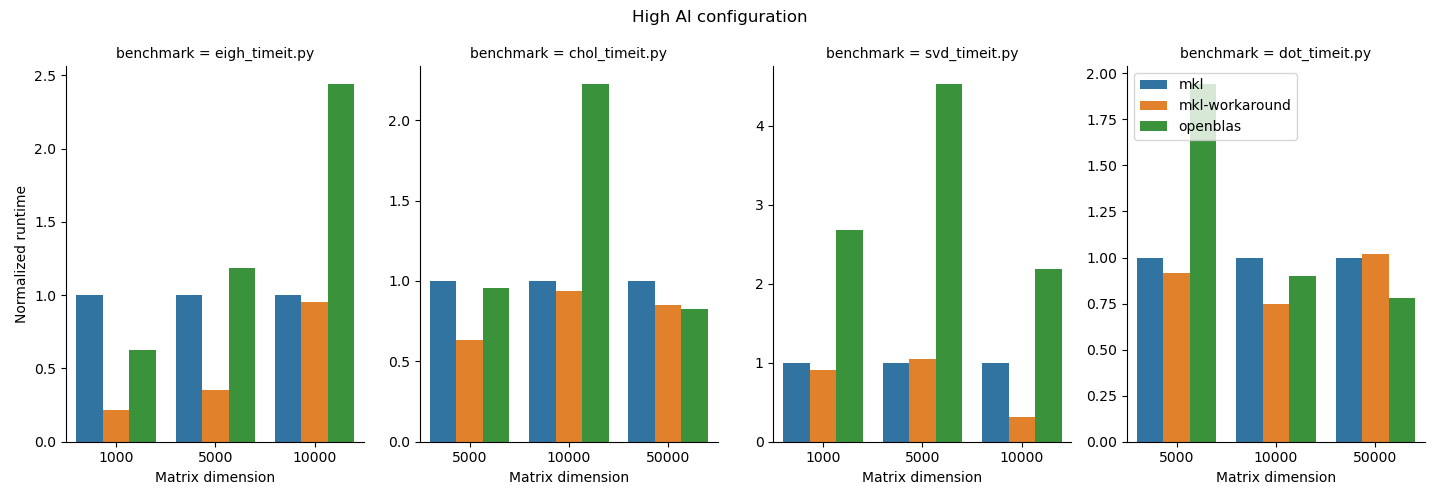
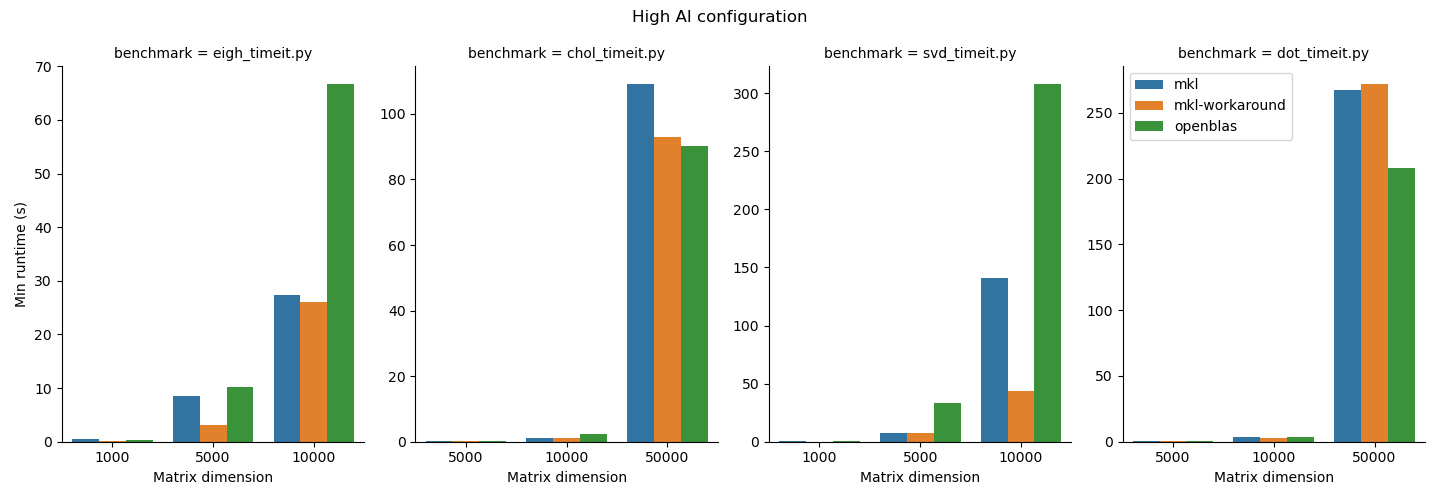
Benchmarking study results¶
- For the functions we studied here, the MKL workaround can add some performance improvements, but it depends on the matrix size and function type. Notably also it added runtime to the
np.dotfunction, which underlines the importance of doing some basic benchmarking. We do suggest that users who need performance test out the MKL workaround since it is straightforward to use. - OpenBLAS beat MKL in
np.linalg.choleskyin thehighAIconfiguration and innp.dotin both configurations. MKL may not always be faster; again, it is important to benchmark your application. 1.np.linalg.eigh,np.linalg.cholesky, andnp.linalg.svdperformed better in thehighMBconfiguration andnp.dotgenerally performed better in thehighAIconfiguration. Python users should be aware that their choice of process and thread binding can also have a large impact on performance. - Other libraries like BLIS exist. BLIS in general was slow in our testing so we did not include it here. BLIS is under active development so we'll keep an eye on this effort and update our reccomendations as the landscape evolves.
- This benchmark was performed on our
/global/common/softwarefilesystem. Python users are advised to remember that other factors influence performance, especially choice of filesystems and containers. Please see this page for tips on improving general Python performance at NERSC.
We summarize the results of the benchmarking study grouped according to highMB and highAI configurations. For each configuration, we show the runtime normalized to mkl on top and the minimum runtime in seconds on the bottom.